What Is The Function Of Restriction Enzymes – When it comes to producing mRNA vaccines, you need to start how you want to finish – and do it very well. It’s the first step in
Transcription (IVT) relies on selecting the best restriction enzyme (RE) to clone the model plasmid. The leading scientific resource for characterization, quality, and manufacturing, New England Biolabs
What Is The Function Of Restriction Enzymes

The COVID-19 pandemic has prompted vaccine development at a record pace, and mRNA-based vaccines have emerged as an alternative to traditional vaccine approaches. mRNA expression is transient and rapid. It’s easy to compile, compare and scale – these are good things because they reinforce many discoveries and early development efforts. Once the viral genome is sequenced, the DNA sequence of interest (which can be used as a vaccine target) can be identified within a few days. An amazing scientific success story.
Ecori Restriction Site
The method of making mRNA vaccines is enzymatic: after the plasmid containing the vaccine target sequence is transformed into bacteria, it is fermented on a large scale and harvested, the workflow uses many enzymes . The plasmid is then sequenced with a restriction enzyme, and the DNA is used as a template for
Transcription to produce mRNA. Additionally, mRNA requires cotranscriptional or enzymatic capping with a poly(A) tail to function. The resulting mRNA is eventually mixed with lipid nanoparticles and injected as a vaccine, thereby producing the antigen required to elicit an immune response. And while the mRNA synthesis process has been around for a long time and is used in many biotechnological applications, synthetic mRNA was shown to generate an immune response in 2012 and was tested as a vaccine against the virus Zika in 2017. The requirements for the enzymes used to make mRNA in pharmaceuticals and produce vaccines are different.
One of the first major steps in developing a SARS-CoV-2 vaccine, for example, is to sequence the plasmid DNA that contains the gene that encodes the viral spike protein. Restriction enzymes are used to ligate the template plasmids. Therefore, it is important to ensure that the production and production of enzymes are carried out in an animal-free environment and adhere to high quality standards. have been involved in the research and development of binding enzymes for almost 50 years – our expertise in this area allows us to provide the following guidelines and things to consider when selecting an enzyme inhibitor for vaccine development:
Animal-derived substances, such as heparin, are often used to purify inhibitory enzymes. Enzyme restriction is processed and manufactured with/without animal derived products.
Dr.amina El –shaibany.
Albumins, such as BSA, help stabilize enzymes when incorporated into final formulations. release final formulation with recombinant Albumin (rAlbumin), ensuring that no animal products are present to support the FDA’s regulatory efforts.
The longer the recognition sequence, the less likely it is that the sequence will appear in the gene fragment and the easier it is to construct a plasmid template. It is therefore advantageous to select inhibitory enzymes with long recognition sequences. send many enzymes to 7-8 base recognition sequence.
Also, don’t leave more than 3′ above the curb. The presence of a 3′ post will increase the production of by-products from the model over time.

Some enzymes are better than others. When choosing an enzyme, make sure you are finding the most stable enzyme possible (24 months shelf life and -20°C storage is ideal). Watch out for enzymes with a shorter shelf life and -80°C storage recommendations*.
Molecular Biology In Medicine
Most enzyme inhibitors recommend an incubation temperature of 37°C. Be aware that some enzymes require other temperatures. This table lists activity at 37°C if you need a specific enzyme for your production.
Some enzymes require little washing, for example, Triton-X-100, to stabilize the enzyme. It is important to review the manufacturing aspects of your desired product so that the cleaning levels do not conflict with local regulations, for example, REACH in Europe.
Type IIS restriction enzymes are the enzyme of choice for cloning plasmids used in vaccine development because they cut outside the signal sequence and ensure the integrity of the poly( A) tail encoded by the manufacturer. By using Type IIS RE, no “scar” remains on the DNA sample after synthesis, and no nucleotides are needed during IVT.
Multi-site enzymes. Some enzymes require more than one site for optimal localization. In most cases, the recommendation is to add the oligo to the reaction and remove it. Learn more here.
Cloning Techniques In Antibody Discovery And Engineering
For more information on enzymes for your IVT process, including our GMP-grade IVT kit for therapeutic RNA production, please contact our Solutions Solutions team. It’s an expert to find the perfect binder for your needs.
* “GMP-grade” is a code word used to describe reagents manufactured in the Rowley facility. The Rowley facility was designed to manufacture reagents under stricter structures and controls to meet product requirements and customer requirements. The reagents manufactured at the Rowley facility are manufactured in accordance with the quality management system standards ISO 9001 and ISO 13485. However, at this time, they do not manufacture or sell products known as Active Pharmaceutical Ingredients ( APIs), nor does it make its products in accordance with all current Best Practices.
Will not rent, sell or transfer your data to any third party for commercial purposes. See our Privacy Policy for details. Check out our Community Guide.

If you have been idle for more than 20 minutes, for your safety you have been logged out. Please login to continue your session.
The Biobrick Approach
Your profile has been mapped to a House, please sign in to complete your profile updates.
To save your cart and view previous orders, sign in to your account. Adding products to your cart without signing in will lose your cart when you sign in or leave the site.
Although every effort has been made to follow the pronunciation style rules, there may be some exceptions. Please refer to the appropriate manual or other sources of information if you have any questions.
The Editors of the Encyclopaedia Encyclopaedia deal with subjects in which they have a wealth of knowledge, either through years of experience working on that subject or through graduate study. They write updates and verify and edit content received from contributors.
Transcription Translation And Replication Notes: Diagrams & Illustrations
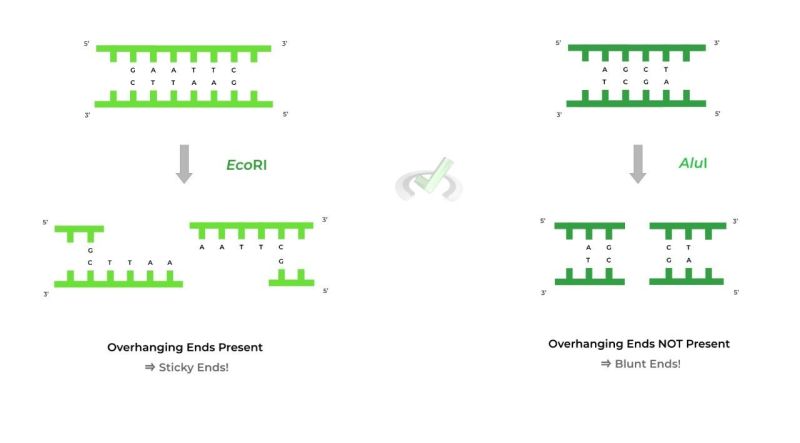
Binding enzyme, a protein produced by bacteria that binds DNA to specific sites along the molecule. Inside the bacterial cell, the inhibitory enzymes cleave the foreign DNA and destroy the pathogens. Restriction enzymes can be isolated from bacterial cells and used in the laboratory to manipulate fragments of DNA, such as those found in genes; for this reason they are essential tools for recombinant DNA technology (genetic engineering).
Bacteria use an inhibitory enzyme to defend against bacterial viruses called bacteriophages or phages. A phage infects a bacterium and inserts its DNA into the bacterial cell so that it can replicate. The restriction enzyme stops the replication of the phage DNA by cutting it into several pieces. Inhibitory enzymes are named for their ability to neutralize or limit the number of strains of bacteriophage that infect the bacteria.
Each binding enzyme recognizes a short, specific sequence of nucleotide bases (the four chemical units of a double-stranded DNA molecule—adenine, cytosine, thymine, and guanine). These regions are called signal sequences, or signal regions, and are normally distributed throughout the DNA. Different types of bacteria produce restriction enzymes that recognize different nucleotide sequences.

When the binding endonuclease recognizes a sequence, it cleaves through the DNA molecule by hydrogenation (the breaking of a chemical bond by adding a water molecule) of the bond between adjacent nucleotides . Bacteria prevent their own DNA from degrading this process by hiding their coding sequence. Enzymes called methylases add methyl groups (—CH
Rnase H Based Analysis Of Synthetic Mrna 5′ Cap Incorporation
) to adenine or cytosine bases in the recognition sequence, which are modified and protected from endonuclease. The restriction enzyme and its methylase correspond to the anti-modification system of a certain type of bacteria.
Historically, four types of binding enzymes have been recognized, designated as I, II, III, and IV, which differ in structure, cleavage site, specificity, and functions. Types I and III are similar in that the inhibitory and methylase activities are performed by one large enzyme complex, unlike the type II system, where the inhibitory enzyme is independent of its methylase. The binding enzymes for type II also differ from types I and III because they target DNA at specific sites within the recognition site; others leave the DNA randomly, sometimes hundreds of bases from the target sequence. Several thousand types of type II enzymes have been identified from various bacterial species. These enzymes recognize a few hundred specific sequences, usually four to eight bases long. Type IV binding enzymes only cleave methylated DNA and show poor sequence alignment.
Inhibitory enzymes were discovered in the late 1960s and early 1970s by Werner Arber, Hamilton O. Smith and Daniel Nathans. The enzymes’ ability to cut the DNA at the right places allowed the researchers to do so