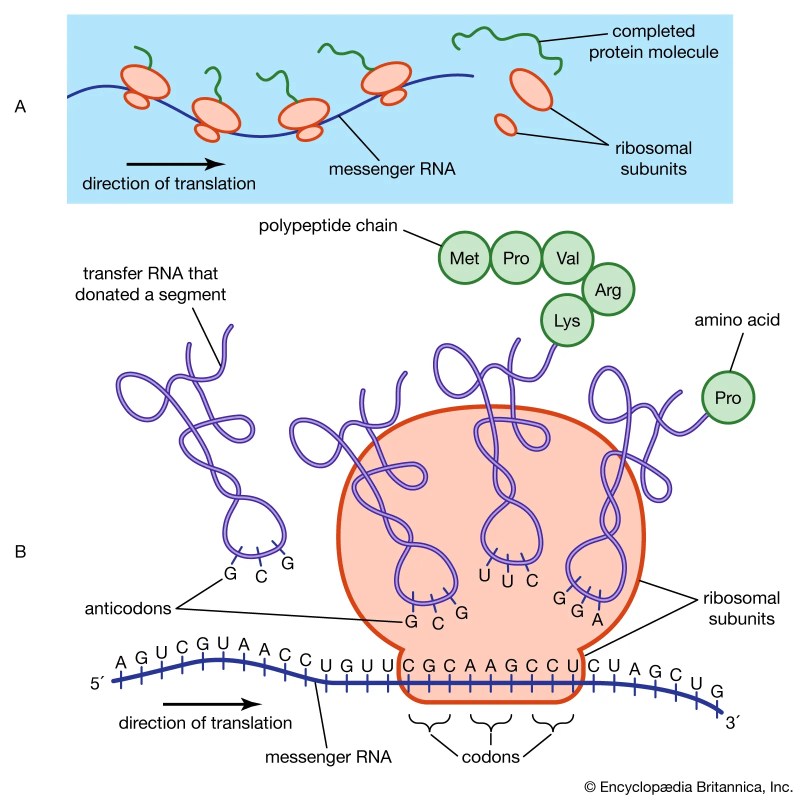
What Happens To Excess Amino Acids In The Body – Accumulating in the liver mitochondria. Ammonia is formed in the liver through oxidative deamination by glutamate dehydrogenase, ending up in urea. Alternatively, it can be added to glutamic acid to form glutamine which can head to the kidney where, after sequential reactions by glutaminase (a deamidation reaction) and glutamate dehydrogenase, it produces ammonia for direct excretion in the urine. If your diet is high in proteins and therefore amino acids, because they cannot be treated as proteins per se, they are metabolized for energy and the metabolic products can be converted into fat or carbohydrate free. It leaves a large amount of nitrogen that is consumed as urea for excretion.
Urea is a water-soluble molecule that cannot be used by chemists in the laboratory to chemically suspend protein (in the range of 3-6 M). Blood urea nitrogen (BUN) values range from 7 to 20 mg/dL N, which is equivalent to 2.5 to 7.1 mM urea. Serum levels are largely dependent on the balance between urea synthesis in the liver and its elimination by the kidney. The amount of normal urea should be the same.
What Happens To Excess Amino Acids In The Body
The color is separated to select the sources of C and N atoms. One N is from ammonia (or from the amido N of glutamine), while the C atom is from carbonate. The other is from the amine of aspartate. Note that 3 ATP are used to power the cycle. See also the guanidino group of arginine (NH(NH
Figure 1 From Metformin
) which readily donates 2 N atoms to the urea-forming step. Knowing that one amino acid readily donates N atoms to urea, you can easily predict that.
Now, let’s look at some of the steps, starting with those used by ATP, which are the ones where nitrogen is incorporated into the precursors of urea.
This enzyme blocks the first “execution” step of the pathway and produces the reactant that enters the cycle, carbamoyl phosphate. It requires a specific cofactor, N-acetyl-glutamate (NAG), which is produced by the acetylation of glutamate by acetyl-CoA using a NAG synthase enzyme activated by arginine. It is not the end of the cycle but provides the input for it.
A cytosolic version of this enzyme, CPS II, is used to synthesize arginine and pyrimidine nucleotides using glutamine as a donor of NH.
Liver, Excretion & Homeostasis
Union for urea by condensing bicarbonate and ammonia to produce carbama, which has a high energy “motif” related to its hydrolysis product. (Note: this does not mean that the bond that is broken is high energy, a misnomer that appears in many books.) So the reaction is powered by 2 ATP as an energy source. Urea is a molecule that “carries” two NH
Both active base molecules (with respect to their hydrolysis products) are protected from nonspecific hydrolysis when the combined anhydride passes through a blocked tunnel to a more distant phosphorylation site of the multimeric enzyme. .
Figure (PageIndex) shows a relative iCn3D model of carbamoyl phosphate synthetase I with bound ADP and N-acetyl-glutamate (5DOU).

Figure (PageIndex): Human carbamoyl phosphate synthetase I with bound ADP and N-acetyl-glutamate (5DOU). (Copyright; original author). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/i…yV8fW3mW9idy6A
Isoelectric Points Of Amino Acids (and How To Calculate Them)
The model represents one chain of the multimer. The 2 ADPs and the single N-acetylglutamate (NLG) are shown as spacefill and labeled. Charged ions are shown (not labeled).
The protein exists in a single monomer-dimer equilibrium. Each monomer contains 1 NAG and 2 ADP, representing two different binding sites for the two phosphorylation steps as shown in the answer above. The two phosphorylation stages are connected by a tunnel that allows the fused anhydride to pass to the carbamate phosphorylation site. NAG is an allosteric modifier because it does not bind to any of the phosphorylation sites. Upon binding to the ring structure of the enzyme, it undergoes a large conformational change that allows the ready enzyme conformation with a full tunnel to prevail.
A diagram of the tunnel for one monomer of carbamoyl phosphate synthetase I is shown in Figure (PageIndex).
The two ADPs below are shown in spacefill with CPK color. The tunnel between the two ADP sites is shown in magenta. The yellow molecule is NAG, which reuses its healing effect by binding to an allosteric site, which effectively opens the tunnel.
Telugu Solution] Excess Amino Acids In The Body Are Broken Down To Fo
Figure (PageIndex) shows the enzyme monomer in a different shape with a tunnel created with ChExVis: a tool for translation and visualization. The large red and green circles represent open spaces.
The top panel in Figure (PageIndex) shows the radius in Angstroms of the tunnel that goes from the red circle to the green circle in the image above. The bottom panel shows the measure of hydrophobicity (red)/hydrophilicity (blue) of the amino acids surrounding the tunnel.
Some ATP is released to convert citrulline to arginosuccinate, as shown in Figure (PageIndex). The product can be separated in the next step to make the amino acid arginine.

This enzyme catalyzes the beta-elimination reaction, which can go through the E2 or E1 mechanism. A simple but very brief process showing the completion of two steps (E1) passing through the carbanion intermediate is shown in Figure (PageIndex): below.
Best Bcaa Supplements: A Dietitian’s Picks
Each of the four monomers is shown in a different color. 3 arginosuccinates (substrate) and 1 fumarate and 1 arginine (product) are shown in spacefill, with CPK colors and highlighted. Each monomer has an N-, M-, and a C center. The four binding sites consist of residues from three monomers. The products are on the fourth level. Catalytic residues may contain a serine and a histidine that act as basic acids/bases with a deprotonated lysine making serine the basic base.
The answering machine is very simple. Carbamoyl phosphate is an activated electrophile in which the carbonyl C is attacked by a deprotonated amino of ornithine to form citrulline. The gene is found on the X chromosome in humans so a mutation in males leads to a “hyperammonemic coma” and death. Heterozygous females can be asymptomatic and this usually results in death. Heterozygous women are asymptomatic or have problems arising from defects in pyrimidine synthesis that can be relieved by a low-protein diet with the addition of arginine.
There are two variants of this enzyme, Arginase I is a cytosolic enzyme expressed in the liver and involved in urea synthesis. Arginase II is a mitochondrial enzyme and is mainly involved in arginine metabolism. Arginase I converts arginine to ornithine and urea. The enzyme contains 2 Mn ions in the active phase. The hydroxide ion binds to two Mn ions and acts as a nucleophile that attacks the guanidino group of arginine and forms a tetrahedral intermediate. Asp 128 acts as a basic acid and removes the amino group to form ornithine and urea. Water recombines with the dinuclear Mn group and dissolves it, donating a proton to the solvent through an intermediary His 41 in the regulation of the active enzyme.
The model below shows the trimeric human arginase I (3KV2) in complex with the inhibitor N(omega)-hydroxy-nor-L-arginine, where one of the guanidino Ns is replaced with an OH. The two gray circles are Mn ions.
News About Amino Acid Metabolism In Plant–microbe Interactions: Trends In Biochemical Sciences
Figure (PageIndex) shows a simulated iCn3D model of trimeric human arginase I in complex with the inhibitor N(omega)-hydroxy-nor-L-arginine (3KV2).
Figure (PageIndex): Trimeric human arginase I in complex with the inhibitor N(omega)-hydroxy-nor-L-arginine (3KV2).(Copyright; original author). Click the image for a popup or use this external link: https://structure.ncbi.nlm.nih.gov/i…QaAQpt5t2KKic6
Water molecules can act as ligands and bind transition state metal ions through covalent bonding. The pKa of the bound water changes to a lower value, making it more likely to deprotonate and become OH

, which is a more efficient base and nucleophile. Water 480 in the active site forms a coordinate covalent bond to the Mn ion creating a strong hydroxide base to attack the guanidino group of arginine, leading to the formation of urea and ornithine.
Solution: Excretion Part 1 Biology Igcse Handwritten Notes All Chapters Covered
If too many amino acids/proteins are depleted, changes in gene expression can increase the levels of enzymes in the urea cycle. In the short term, the activity is regulated by the enzyme that provides access to the cycle, carbamoyl phosphate synthase, whose activity is regulated by N-acetylglutamic acid and which separates the limiting step . As mentioned above, without the allosteric regulator, NAG, the enzyme has no activity. The levels of NAG are determined by the enzyme NAG synthase, which is regulated by free arginine, the precursor of urea in the cycle. Supplemental arginine is given to patients with urea cycle disease and may have an effect through the regulation of NAG synthase.
Two of the major pathways you explored, the TCA and urea cycles, are not linear but circular. What advantages does “circular” have over linear ones? A comparison of economic means provides an insight. In a linear economy, raw materials are used to make a product that is discarded as waste. Unfortunately, linear methods have conquered our world at great cost to our environment. To reduce inefficiencies in line economy and work
Excess amino acids, what happens to excess amino acids in the liver, what happens to excess amino acids, amino acids in body, amino acids in the body, what happens to amino acids in the liver, what happens to excess protein, excess of amino acids in the body, amino acids in human body, in the body excess amino acids are, body health amino acids, excess amino acids in the body